- Joined
- Mar 18, 2008
- Messages
- 5,717 (0.97/day)
| System Name | Virtual Reality / Bioinformatics |
|---|---|
| Processor | Undead CPU |
| Motherboard | Undead TUF X99 |
| Cooling | Noctua NH-D15 |
| Memory | GSkill 128GB DDR4-3000 |
| Video Card(s) | EVGA RTX 3090 FTW3 Ultra |
| Storage | Samsung 960 Pro 1TB + 860 EVO 2TB + WD Black 5TB |
| Display(s) | 32'' 4K Dell |
| Case | Fractal Design R5 |
| Audio Device(s) | BOSE 2.0 |
| Power Supply | Seasonic 850watt |
| Mouse | Logitech Master MX |
| Keyboard | Corsair K70 Cherry MX Blue |
| VR HMD | HTC Vive + Oculus Quest 2 |
| Software | Windows 10 P |
What else can we use our "gaming GPU" other than gaming? Mining? Protein folding? Now we have another option; decoding DNA sequencing data!
Currently on the DNA sequencing front we have 2 main generations of sequencing technology. Traditional 2nd gen short DNA sequencing which usually sequence DNA in 50 basepairs to 500 base pairs. The software side for this tech is already extremely robust. Major draw back is the short DNA size. Considering our genome is 3 billion basepairs long for one set of our diploid genome, even at 500bp it is a drop in the occean. Not to mention 2nd gen DNA sequencing sucks at repetitive regions.
Enter 3rd gen DNA sequencing, where Pacific Bioscience and Oxford Nanopore shines. Way longer read length (hundreds of thousands of base pairs long versus max at 500~ish).
Here is a short video of how the Oxford side works
In short, sequencing machine turns DNA's A,T,C and G base coding into electric current chromatography. This raw file needs to be decoded into traditional DNA sequence file format FASTQ, where the A,T,C and G are reported along with the quality score of each of the base-pairs.
Nvidia has been helping Oxford Nanopore in accleration of decoding such data dense file format. Such decoding process takes way faster on Nvidia CUDA based GPUs versus CPU approach since the decoding utilizes Nvidia's CUDA acceleration.
In the past, Nvidia and ONT has only developed this pipeline for Linux based system. Recently ONT implemented this process into native Windows 10 CUDA system. Which brings me to this point. I have long wanted to have something for bench marking modern hardware for bioinformatics usage. This implementation would allow such direct comparison to happen at direct consumer level. Since all the related genomic files are publicly available.
So what do you need?
A windows10 system with Nvidia GPU, Pascal and forward.
Latest Nvidia driver, and yes Game driver works!
CUDA dev toolkit 11.4 or whatever latest version there are
As for dataset, I will be using a set of human genomic DNA raw read files from ONT's public database
 github.com
github.com
Specifically I used this dataset:
Downloaded the Fast5 (NOT fastq), inflate it to a local SSD folder (and yes you do need a SSD to eliminate bottleneck)
For the basecaller, you can get it here
Installation of the basecaller can be found here
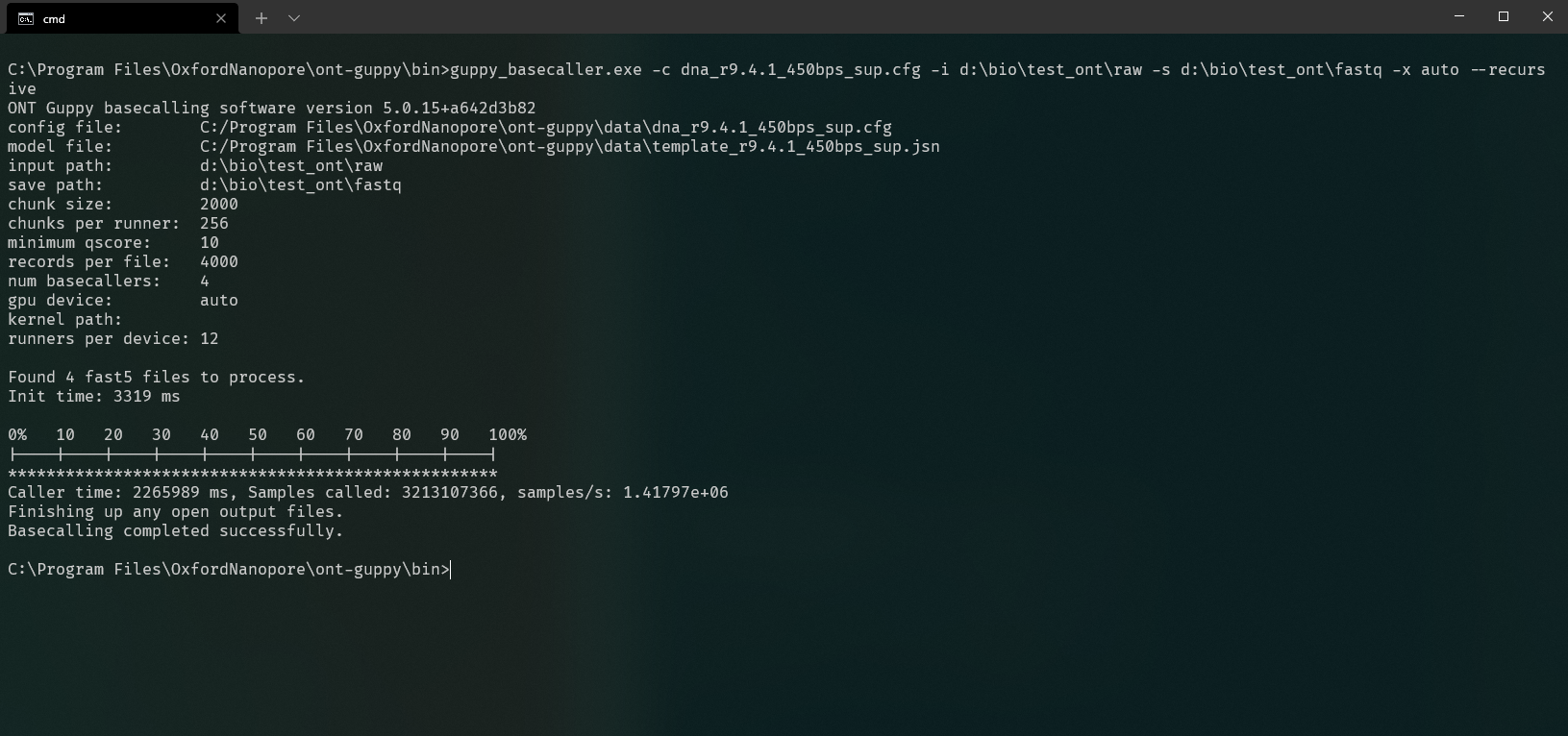
C:\Program Files\OxfordNanopore\ont-guppy\bin>guppy_basecaller.exe
Now just need to point the basecaller to the directory where raw FAST5 data is stored and execute the command
guppy_basecaller.exe -c dna_r9.4.1_450bps_sup.cfg -i <input fast5 path> -s <output fast5 path> -x auto --recursive
Screen will print out the progress. After completion you will get your system speed!
In this case, it is 1.41797e+06 sample per second for the RTX 3090 running max overclock I can get
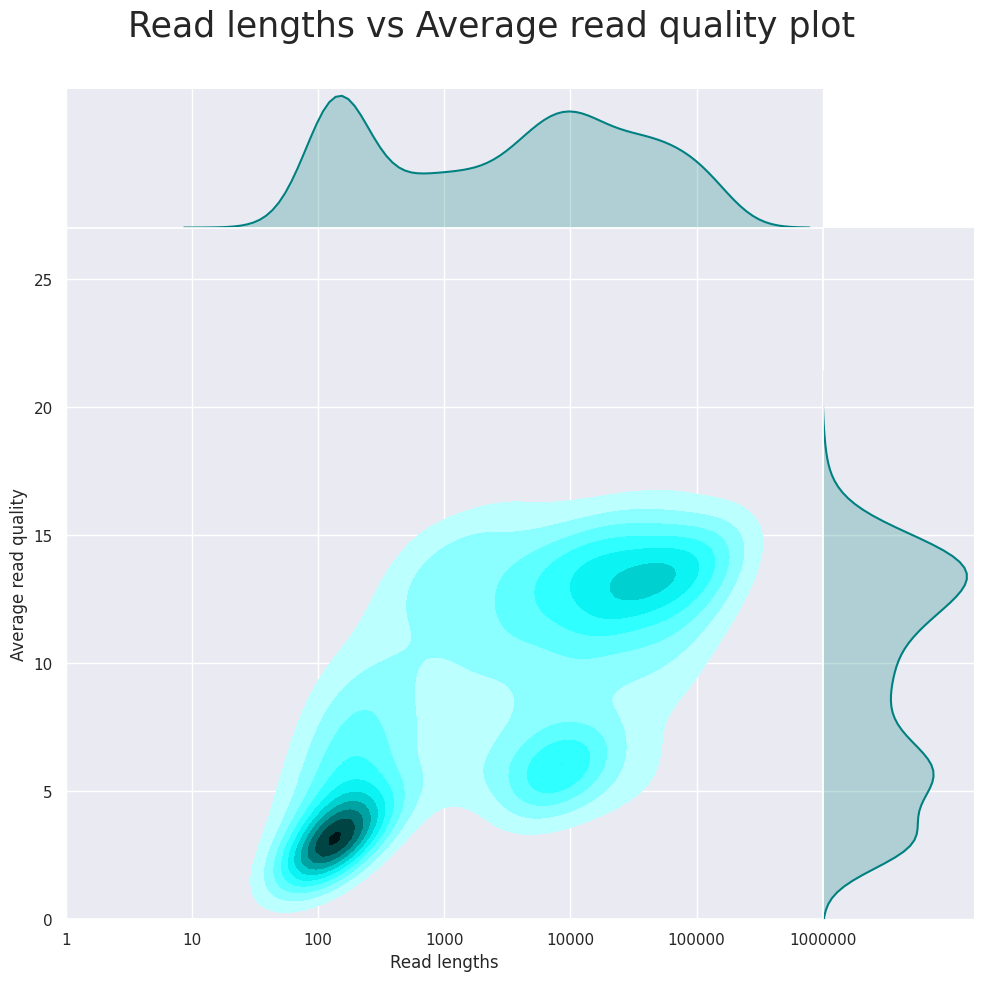
Read quality is OK, as this is a test data set specifically designed for super long DNA reads
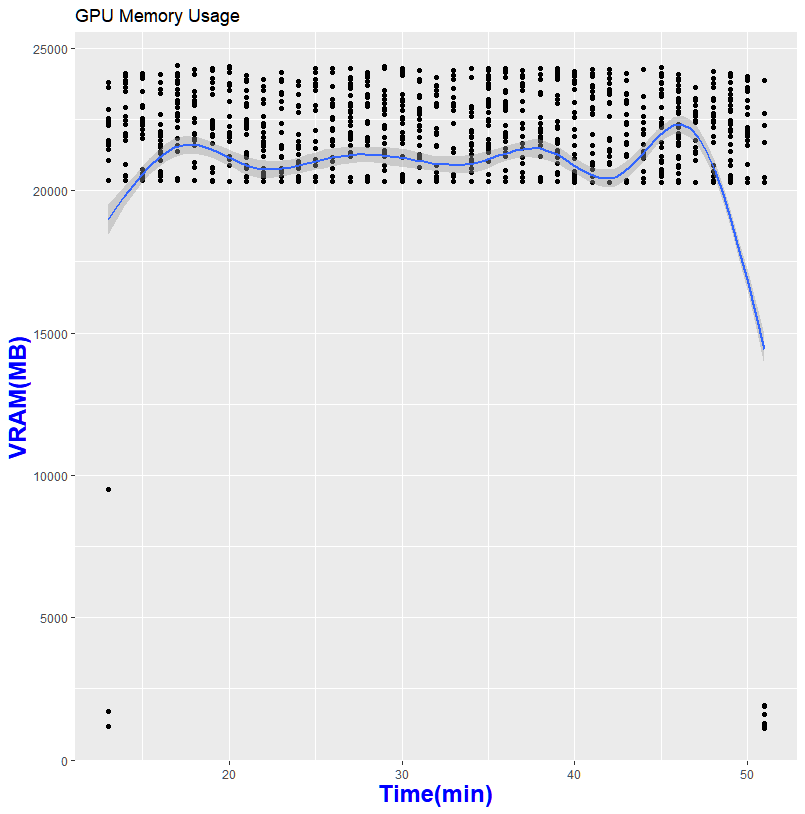
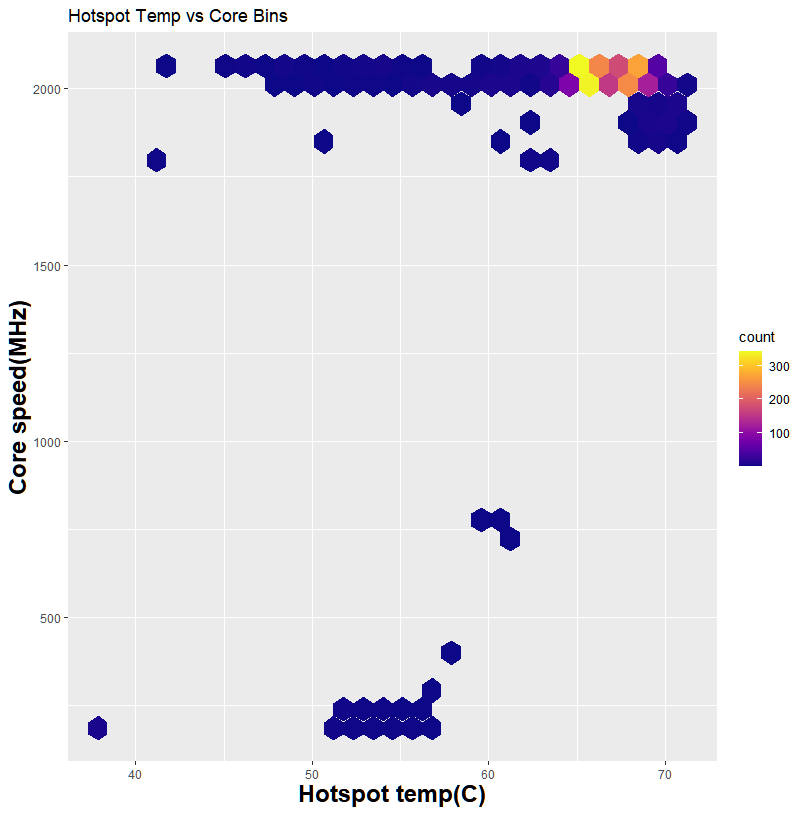
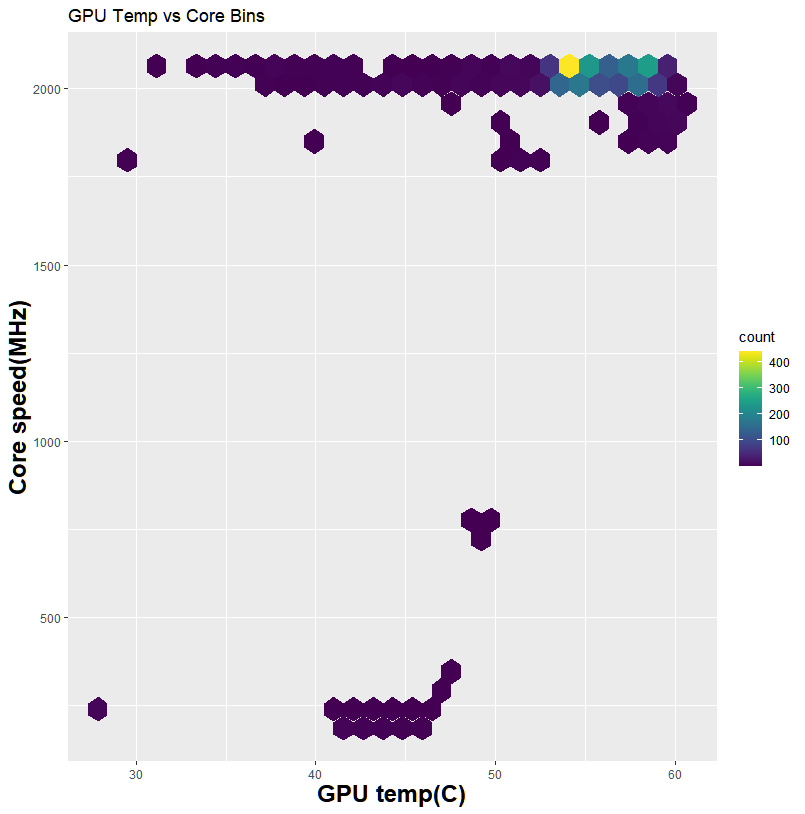
My system during the run. The load is the whole system from CPU to RAM and GPU. Larger datasets can see up to 40GB of system memory usage
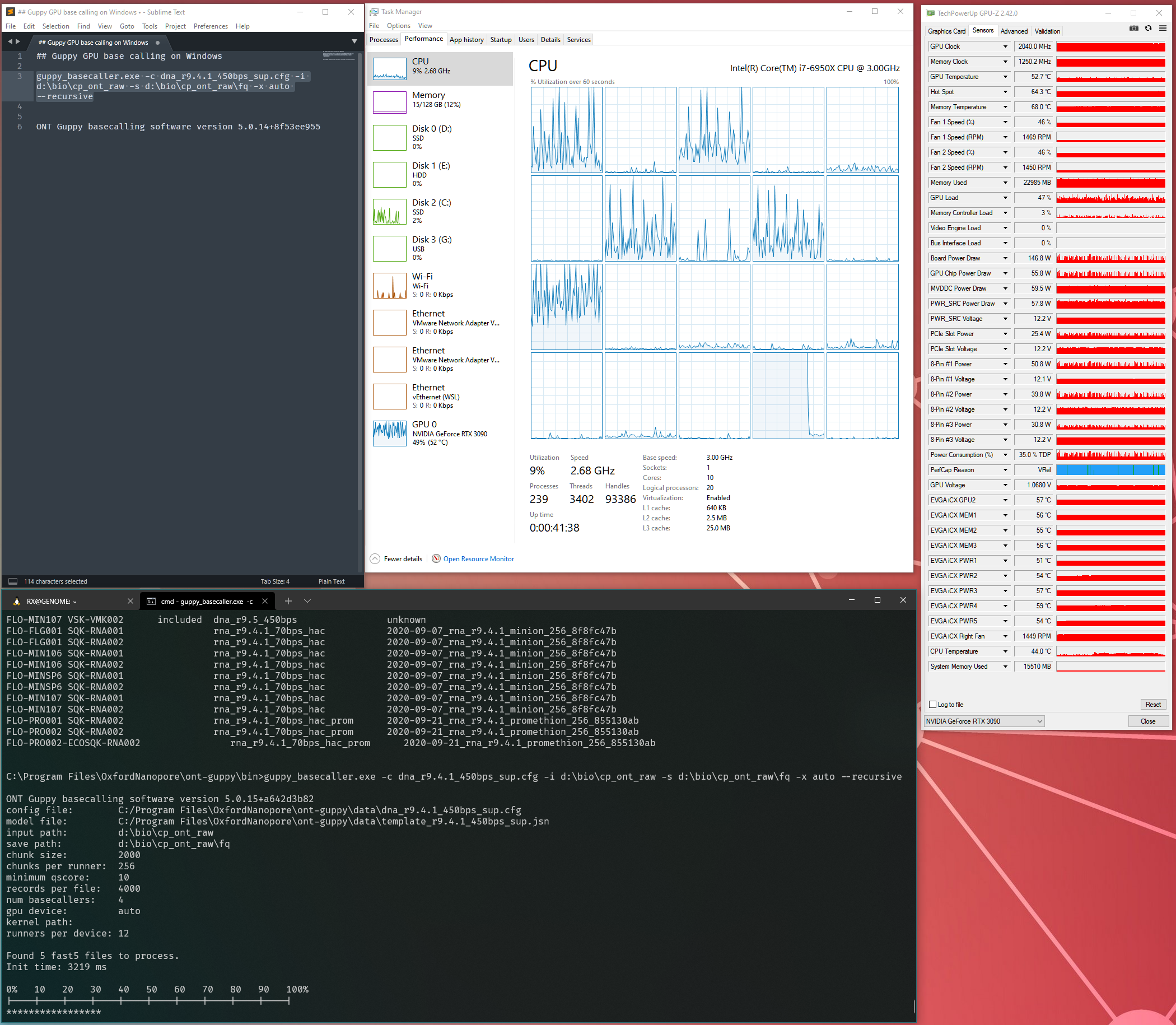
Thanks to GPU-Z, I can log what is happening during my run
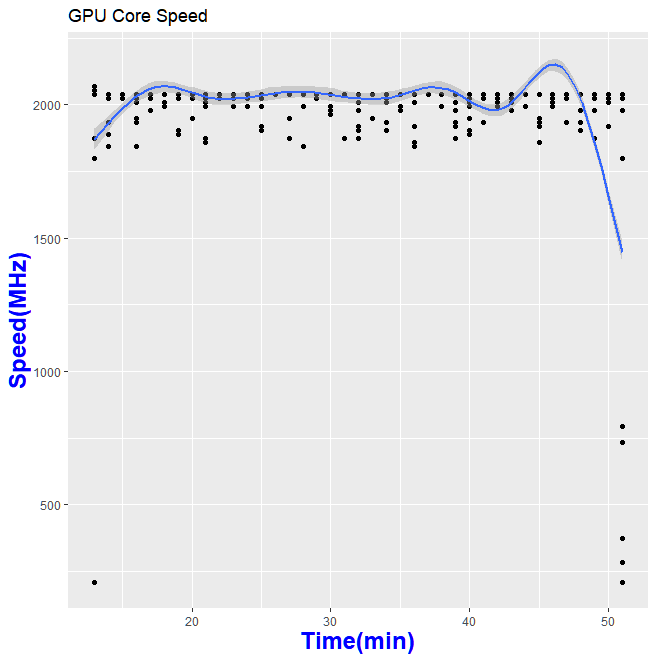
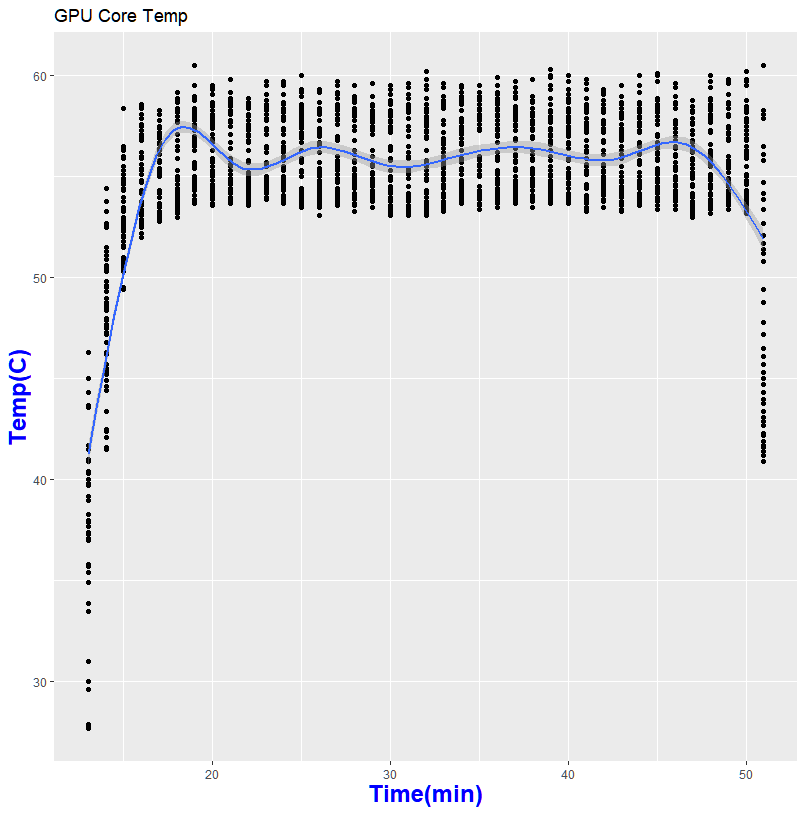
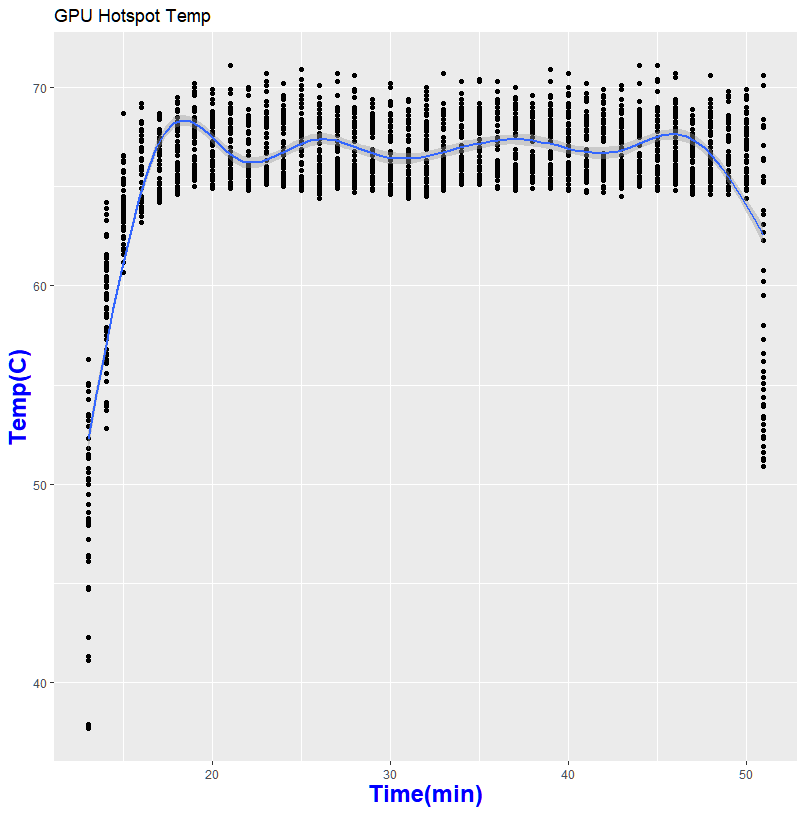
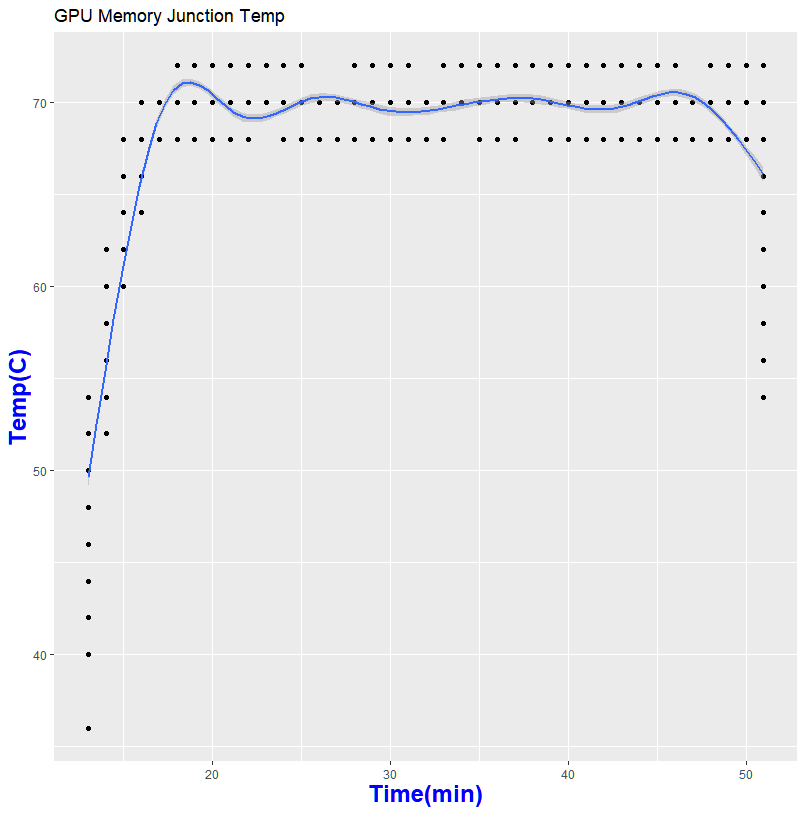
So what does this mean in for folks who wanted to help out science?
In addition to Fold@home looking at GPU docking. Maybe we can come with some distributed computing platform that allow end user to help processing vast amount of DNA / RNA sequencing data.
Well, you can totally benchmark your system / GPU and see how good it is at DNA sequencing processing.
Currently on the DNA sequencing front we have 2 main generations of sequencing technology. Traditional 2nd gen short DNA sequencing which usually sequence DNA in 50 basepairs to 500 base pairs. The software side for this tech is already extremely robust. Major draw back is the short DNA size. Considering our genome is 3 billion basepairs long for one set of our diploid genome, even at 500bp it is a drop in the occean. Not to mention 2nd gen DNA sequencing sucks at repetitive regions.
Enter 3rd gen DNA sequencing, where Pacific Bioscience and Oxford Nanopore shines. Way longer read length (hundreds of thousands of base pairs long versus max at 500~ish).
Here is a short video of how the Oxford side works
In short, sequencing machine turns DNA's A,T,C and G base coding into electric current chromatography. This raw file needs to be decoded into traditional DNA sequence file format FASTQ, where the A,T,C and G are reported along with the quality score of each of the base-pairs.
Nvidia has been helping Oxford Nanopore in accleration of decoding such data dense file format. Such decoding process takes way faster on Nvidia CUDA based GPUs versus CPU approach since the decoding utilizes Nvidia's CUDA acceleration.
In the past, Nvidia and ONT has only developed this pipeline for Linux based system. Recently ONT implemented this process into native Windows 10 CUDA system. Which brings me to this point. I have long wanted to have something for bench marking modern hardware for bioinformatics usage. This implementation would allow such direct comparison to happen at direct consumer level. Since all the related genomic files are publicly available.
So what do you need?
A windows10 system with Nvidia GPU, Pascal and forward.
Latest Nvidia driver, and yes Game driver works!
CUDA dev toolkit 11.4 or whatever latest version there are
As for dataset, I will be using a set of human genomic DNA raw read files from ONT's public database
NA12878/Genome.md at master · nanopore-wgs-consortium/NA12878
Data and analysis for NA12878 genome on nanopore. Contribute to nanopore-wgs-consortium/NA12878 development by creating an account on GitHub.
Specifically I used this dataset:
| FAF15630 | 4244782843 |
Downloaded the Fast5 (NOT fastq), inflate it to a local SSD folder (and yes you do need a SSD to eliminate bottleneck)
For the basecaller, you can get it here
Installation of the basecaller can be found here
C:\Program Files\OxfordNanopore\ont-guppy\bin>guppy_basecaller.exe
Now just need to point the basecaller to the directory where raw FAST5 data is stored and execute the command
guppy_basecaller.exe -c dna_r9.4.1_450bps_sup.cfg -i <input fast5 path> -s <output fast5 path> -x auto --recursive
Screen will print out the progress. After completion you will get your system speed!
In this case, it is 1.41797e+06 sample per second for the RTX 3090 running max overclock I can get
Read quality is OK, as this is a test data set specifically designed for super long DNA reads
My system during the run. The load is the whole system from CPU to RAM and GPU. Larger datasets can see up to 40GB of system memory usage
Thanks to GPU-Z, I can log what is happening during my run
So what does this mean in for folks who wanted to help out science?
In addition to Fold@home looking at GPU docking. Maybe we can come with some distributed computing platform that allow end user to help processing vast amount of DNA / RNA sequencing data.
Well, you can totally benchmark your system / GPU and see how good it is at DNA sequencing processing.

